Research
Hello there! My name is Dongyan (pronounced Dong-yan), I am a researcher in the Fundamental AI Research (FAIR) team at Meta in NYC, working on building AI models that think and act more human-like. Previously I did my PhD at Integrated Program in Neuroscience at McGill University and Mila, supervised by Blake Richards. My research interest lies at the intersection of artificial intelligence and neuroscience. Specifically, I am interested in unraveling the general principles that govern both biological and artificial intelligence, through the lens of representation learning.
Projects
Temporal encoding in Deep Reinforcement Learning agents
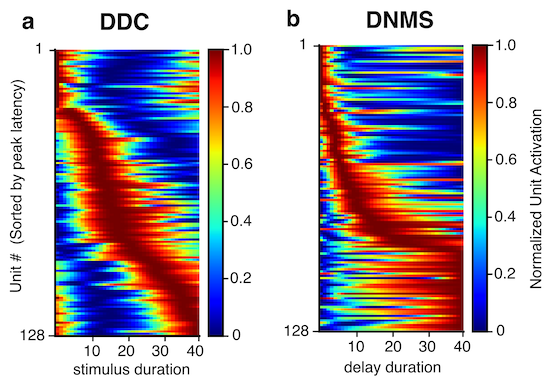
To what extent do the temporal representations in the brain contribute to behaviour? Here, we trained deep reinforcement learning agents on
timing and time-dependent tasks, and showed the emergence of brain-like time representations in networks trained to calculate time implicitly. With virtual lesion experiments, we showed that the temporal representations largely contribute
to behaviour through recurrent dynamics, demonstrating dissociation between temporal tuning curves and timing behaviour.
(Lin et al., 2023 Sci Rep / Lin & Richards, 2021 bioRxiv)
Deep-learning generated optimized images reveal tuning landscape in mouse higher visual cortex

In this work, we set out to answer the questions
"what do neurons in the visual cortex like to see? Do neurons in different visual areas like to see different images?" with deep learning models.
We show that mouse higher visual areas indeed prefer different visual stimuli, but their tuning landscape is more complex than single-neuron preference!
(Preprint / SfN 2022)
Reconciling the Sherringtonian and Hopfieldian views on neural computations
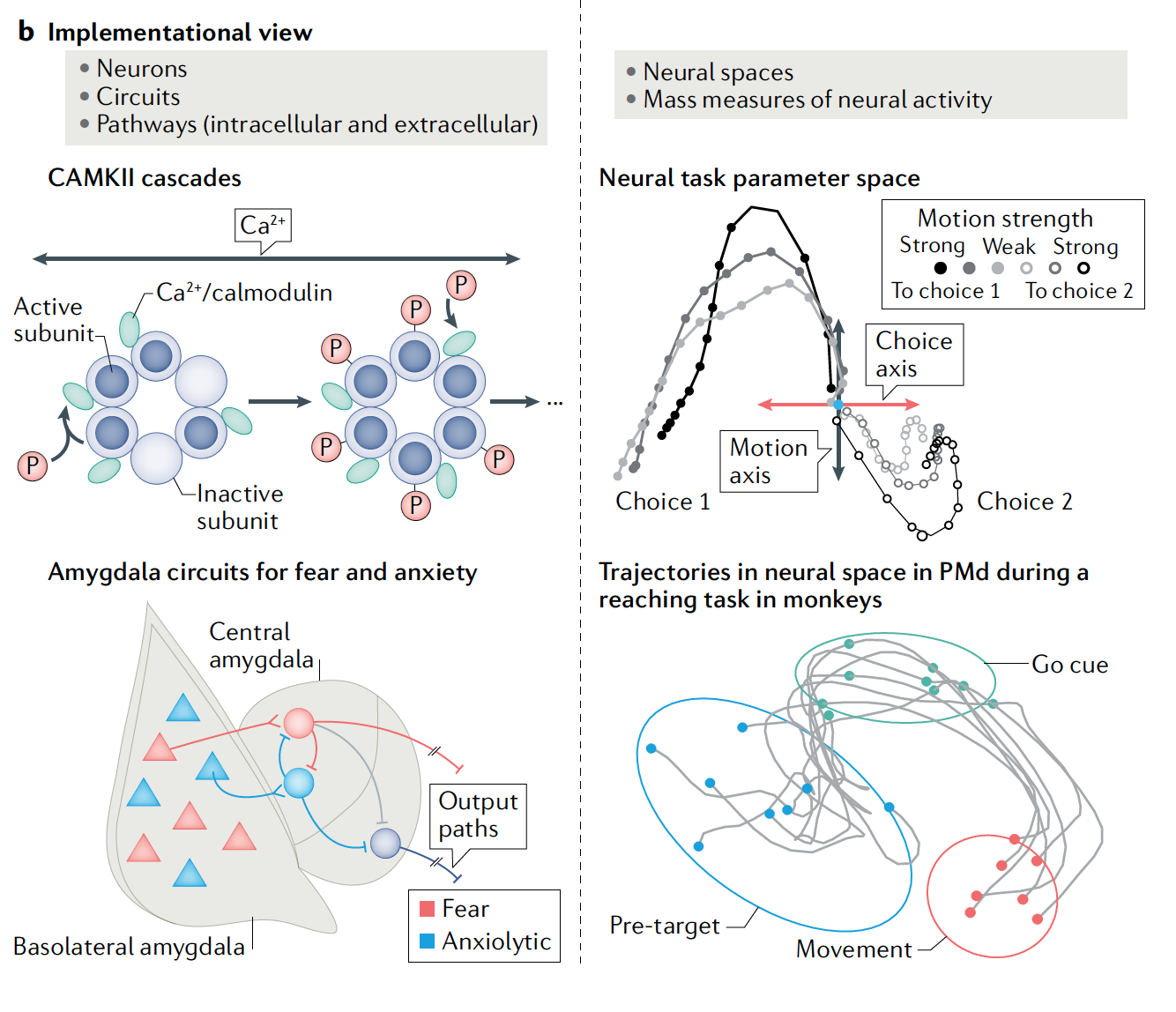
Since the original paper from Barack and Krakauer (2021) came out, the field of
cognitive computational neuroscience has been debating if the brain is better understood from a single-neuron ("Sherringtonian") or population geometry ("Hopfieldian") approach, or if they could be merged, or if they should remain distinct but complementary perspectives.
We organized this GAC workshop to bring together researchers whose work provides different perspectives on the two paradigms, and the possibility of their reconciliation.
(CCN 2023 workshop recording / Proposal)
Deep sequencing of short capped RNAs reveals novel families of noncoding RNAs
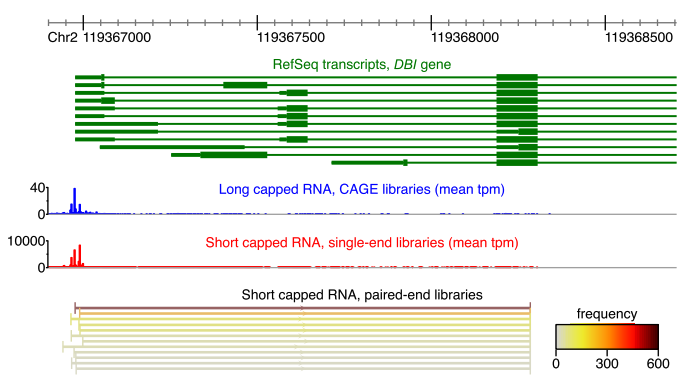
I'm grateful to have contributed to the work through my internship at RIKEN. We developed a sequencing protocol for short capped RNAs, applied it to the human cell line THP-1, and compared it with the landscape of long capped RNAs,
discovering distinct transcription initiation preferences and associations with disease SNPs. This deep sequencing elucidates the diverse array of transcripts from known and novel promoters and enhancers, revealing new noncoding RNA families.
(de Hoon et al., 2022 Genome Research)
Personal
The purpose of this page is to share with you some of the things that I enjoy outside the lab. (Nov 2023 quick note: I don't update this page often (like, once every 3 years), so it's not the most up-to-date reflection of my taste and preference, which change over time. Feel free to chat with me to see what I'm into currently.)
Music, TV, Movies
Below are some of my go-to albums:
My all-time favourite TV show is Breaking Bad. Honourable mentions include: BoJack Horseman, Friends, The Newsroom, and How I Met Your Mother.
I really like the early works of Quentin Tarantino.
Curriculum Vitae
You can download my CV here.




